Spatial Biology Core
Welcome to the Department of Laboratory Medicine and Pathology (DLMP) Spatial Biology Core facility! We are happy to provide molecular annotation and tissue interrogation services for spatial profiling experiments of RNA and protein in tissue sections on the NanoString GeoMx DSP and CosMx SMI platforms. The facility is located in the E-building of the University of Washington’s South Lake Union (SLU) campus and is staffed by experienced research scientists. The core facility is overseen by pathologists with hands-on experience executing spatial profiling experiments REF, REF. using spatial profiling on human tissues. The core facility is located adjacent to the Pathology Research Services laboratory enabling seamless data generation from submitted tissues/tissue blocks as well as downstream validation of spatial profiling experiments' results using in situ hybridization (ISH) and immunohistochemistry (IHC).
Director: Shreeram Akilesh, MD PhD
Co-Director: Kelly D. Smith, MD PhD
Research Scientists: Emily Beirne, BS HTL(ASCP) and James Li, PhD HTL(ASCP)
Why Perform Spatial Profiling?
Spatially resolved profiling of tissues was Nature Methods’ 2020 Method of the Year. Bulk profiling approaches such as RNA-seq lose information about the relative composition of the tissue. Even techniques which excel at identifying rare cell populations such as single cell RNA-seq, lose information about where in the tissue those cells are located. Spatial profiling enables deep phenotypic (protein/RNA) profiling and mapping of those profiles to specific locations within the tissue context. This is especially powerful when the processes under study are unevenly distributed in the tissue (e.g., patchy disease involvement). Recent technological innovations have improved resolution to the single-cell level. Highly-multiplexed (>50 and sometimes >500) detection of proteins is also possible with these platforms. And all of these can be performed on a single 5µm tissue section of archival tissue (FFPE)!
Platforms
The Spatial Biology Core currently has two different platforms that enable spatially resolved profiling. These are the GeoMx Digital Spatial Profiler (GeoMx DSP) and the CosMx Spatial Molecular Imager (CosMx SMI), both from NanoString, Inc. The choice of which platform to use is guided by the experimental question at hand and budgetary constraints. The conceptual basis of these platforms is explained below and the Core's scientists are available in helping you guide selection of one or the other platform for your experiment. We are aware of Nanostring's financial restructuring process and are in close communications with their representatives. We have seen no interruption in client-facing functions such as reagent orders and technical support. Therefore, all experimental planning and execution is proceeding as normal.
GeoMx DSP
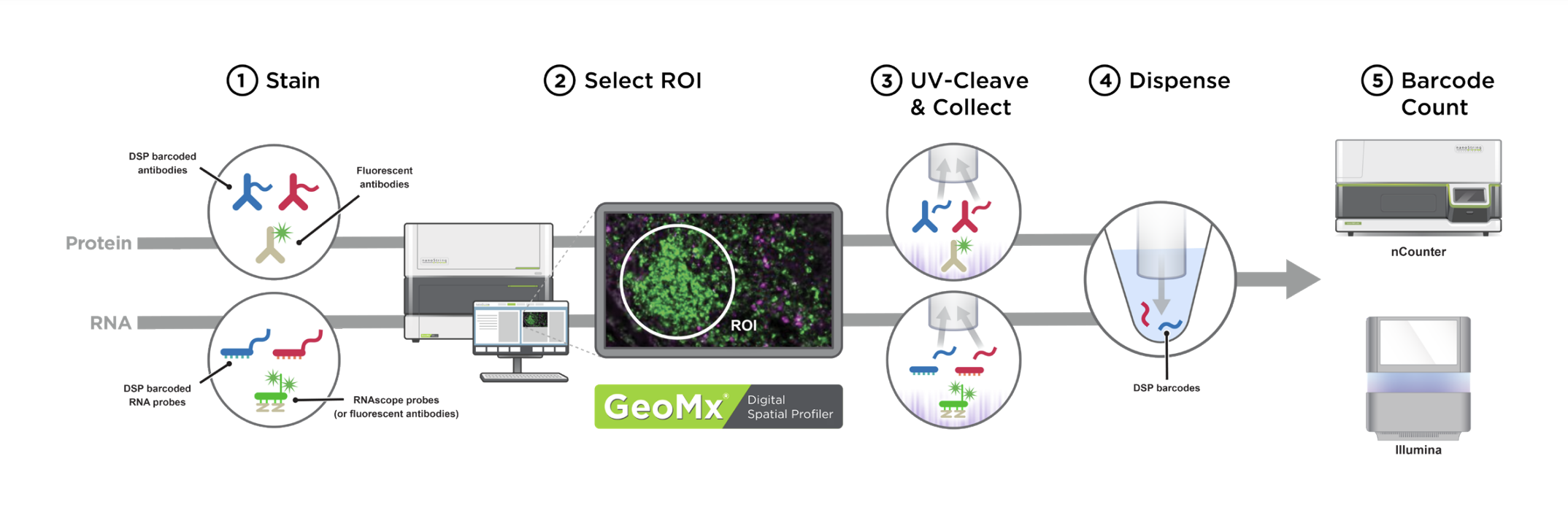
Image detailing the five phases of the DSP process.
Click image to enlarge. Source: GeoMx DSP Grant Support Package.
The NanoString GeoMx DSP permits direct quantification of proteins or RNA transcripts to spatially resolved anatomic structures in tissue sections. After hybridization of specially oligo-conjugated antibodies or RNA in situ hybridization (RNA-ISH) probes to a section, the tissue is counterstained and imaged using immunofluorescence microscopy. Structures of interest are visualized and selected as Regions of Interest (ROI). An ultraviolet laser pulse is focused precisely on this ROI to release oligos conjugated to the target proteins or RNAs. The DSP instrument aspirates these oligos into collection wells using a capillary micropipette. Since these oligos contain barcodes which map them to their source protein/RNA target, digital counting of these oligos with nCounter or next generation sequencing permits a direct readout of target abundance localized to the specific ROI within the tissue section. Read the original paper describing the technology. Read publications using the GeoMx DSP.
CosMx SMI
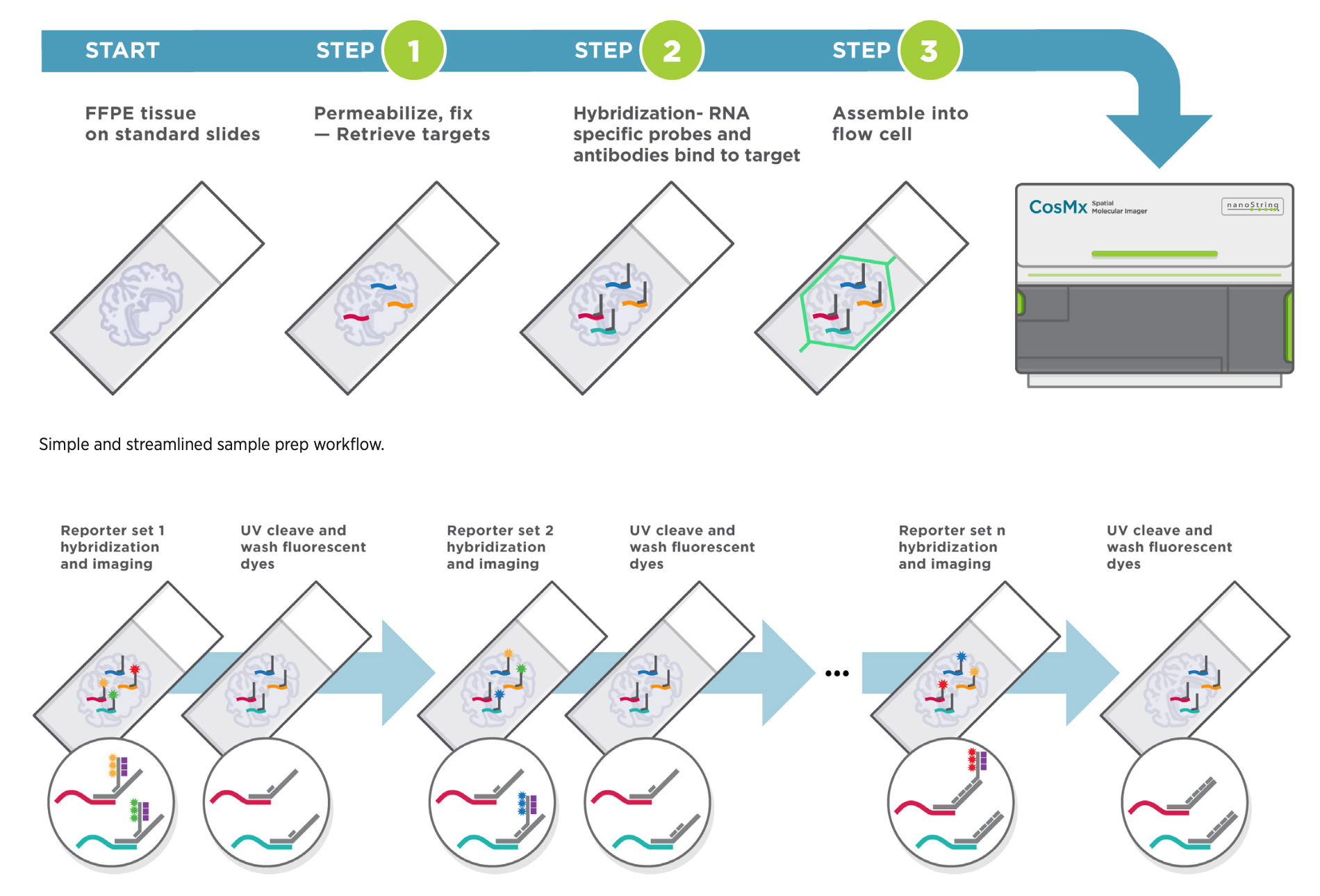
Image detailing the phases of the CosMx SMI process.
Click image to enlarge. Source: CosMx SMI Grant Support Package.
The NanoString CosMx SMI enables single cell resolution quantification of genes or proteins in tissue sections. After hybridization of oligo-conjugated antibodies or RNA in situ hybridization (RNA-ISH) probes to a section, the tissue is exposed to oligonucleotide-based fluorescent readout probes and imaged. Subsequently, bound fluorescent reported are removed and the next round of fluorescent reporters is applied to the tissue. Each target is encoded by a particular combination of reporter oligonucleotides, which enables definitive identification over several rounds of imaging. Cell membrane dyes allow for identification of cell boundaries to assign transcript/protein spots to cells. All data is pushed to the AtoMx cloud platform where additional QC and analysis pipelines can be implemented. Read the original paper describing the technology. More complex analyses such as receptor-ligand interaction and neighborhood analyses can subsequently be performed, as shown for one of our experiments below. For quantification of gene expression, 1000- and 6000-plex panels are available for human tissues (1000-plex mouse panel coming soon). Recently, feasibility of whole transcriptome quantification at single cell resolution has been demonstrated.
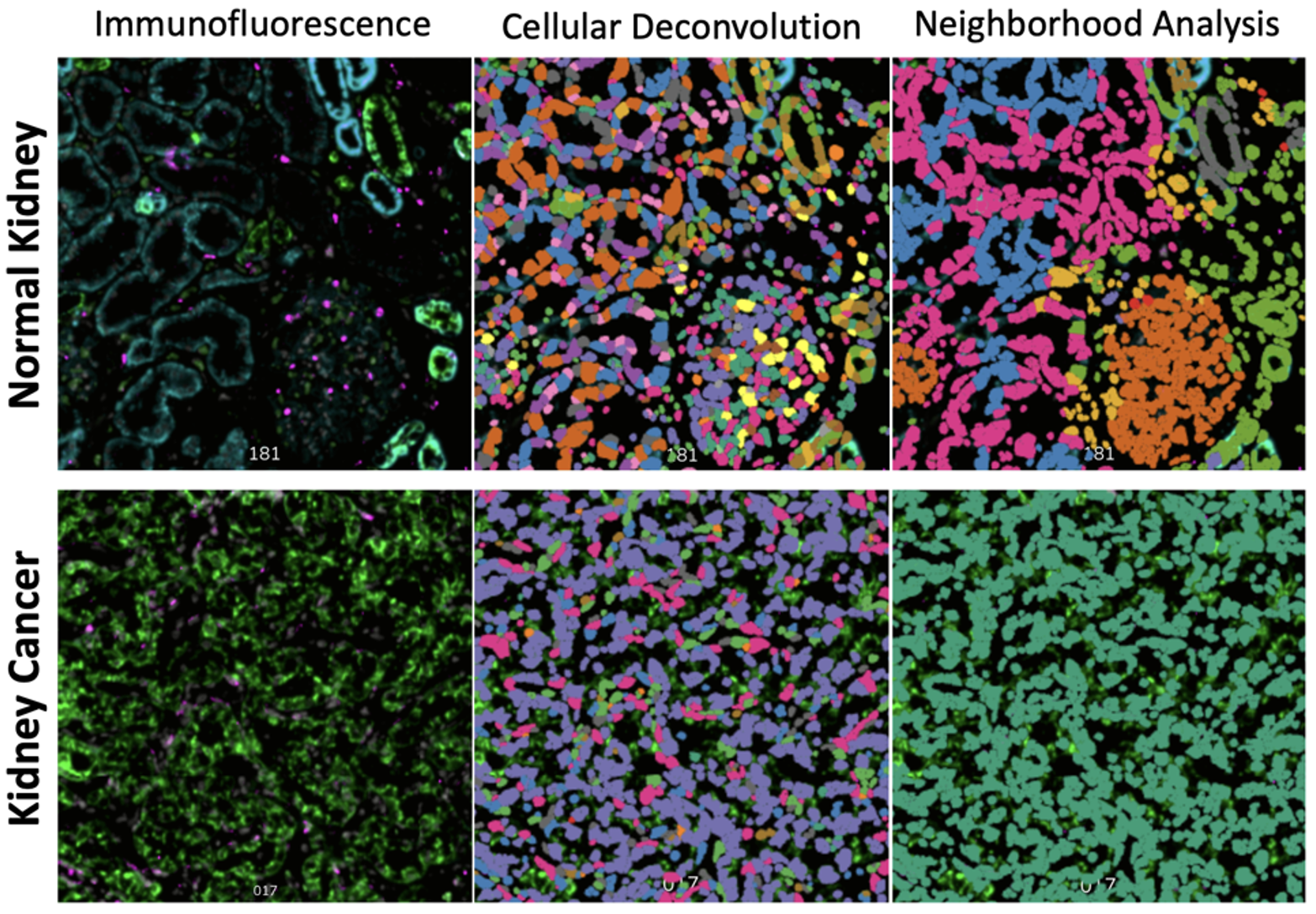
Example of the Core's 1000-plex gene expression profiling with single-cell resolution on the CosMx SMI.
Sample Requirements
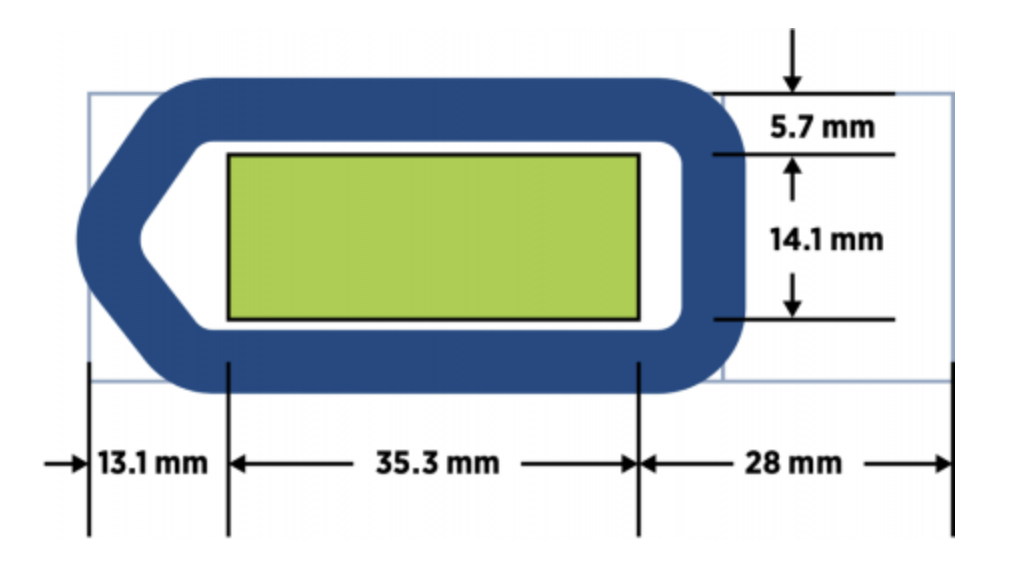
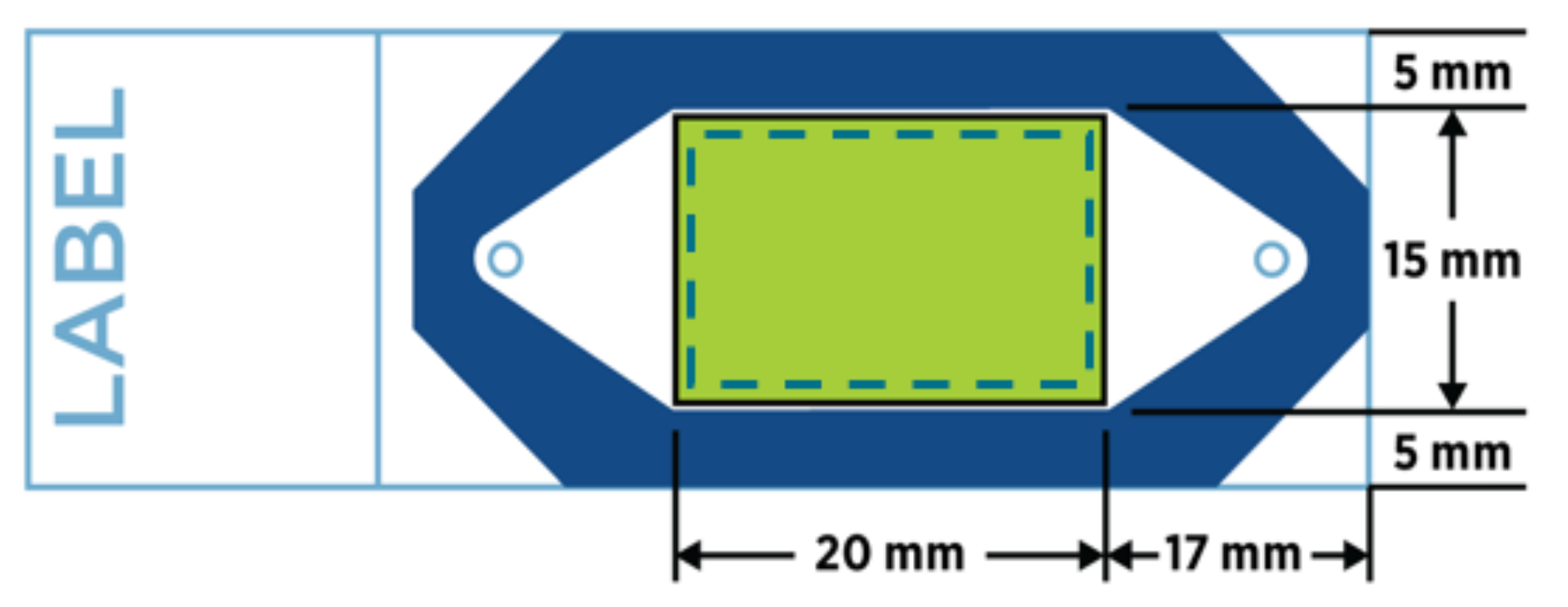
Only a single 4-6µm tissue section (fresh frozen and embedded in OCT or formalin fixed paraffin embedded, FFPE) is required for data generation. If tissues are small, multiple tissues can be placed onto a single slide to reduce reagent costs and to achieve enough replicates for experiment interpretation (e.g., a tissue microarray). Samples must be placed within the green boxed area on a positively charged slide (see schematics below for GeoMx DSP and CosMx SMI experiments). Tissue less than three years old is ideal, but older tissues have also been evaluated successfully. We recommend cutting tissue sections fresh for RNA analysis (within two weeks of the planned experiment; store sections at 4°C with desiccant). For protein assays, sections are stable at room temperature. We strongly recommend coordinating with us prior to cutting your tissue sections. Our staff has extensive sectioning experience and can assist in complex tissue placement with the capture areas. Placing multiple tissue sections within the capture area reduces overall assay costs.
Optimal sample placement for GeoMx DSP.
Optimal sample placement for CosMx SMI.
Source: NanoString GeoMx and CosMx manuals
Getting Started
Since spatial profiling experiments are complex and expensive, we require an initial consultation so that we may assist you with planning and design of your experiment. Please contact us to schedule your consultation. During this consultation, we will discuss your project goals, sample type, desired analyte (protein/RNA), timeline and estimated costs. The major determinants of overall project cost will be the number of desired slides to be processed, region of interest (ROI) number and size, and nCounter/sequencing requirements. As fellow scientists, we are committed to working with you to execute your spatial profiling experiments efficiently, cost-effectively and most importantly, successfully. We can also assist with antibody validation for immunohistochemistry and RNA-ISH for validation of findings obtained through these spatial biology platforms.
Grant Resources
Do you want to propose using the GeoMx DSP or CosMx in your grant? Please find an example grant support package for GeoMx DSP or CosMx. A sample Facilities, Equipment and Resources document for the UW DLMP Spatial Biology Core facility is available here - but please contact us for the most up-to-date version.